In this page, we are dealing with the organization of chromatin during the cell cycle and the morphological features of chromosomes. Genetic information of cells is stored and coded in very large strands of DNA (excepting the much shorter mitochondrial and plastid DNA). DNA is a relatively inert molecule that needs a great deal of help from other molecules, mostly proteins, for getting expressed, regulate such expression, being replicated and for a proper organization in the nucleoplasm. Proteins are also involved during mitosis, where replicated DNA is equally split into two daughter cells. Thus, DNA is always associated to proteins and other molecules. DNA plus associated molecules are known as chromatin.
Cells in interphase (G1, S and G2 phases of the cell cycle) and non-dividing cells show nuclei with heterochromatin, or condensed chromatin, and regions with euchromatin, or loosely condense chromatin (Figure 1). In M phase the whole chromatin is highly compacted in chromosomes. After M phase, chromosomes become both euchromatin and heterochromatin again. Thus, cell division leads to a cycle a condensation and decondensation. During interphase some regions of euchromatin may become condense as heterochromatin and the opposite also happens. These regions alternating between heterochromatin and euchromatin are known as facultative heterochromatin.
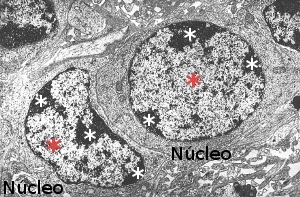
Figura 1. Electron microscopy image showing nuclei with hetherocromatin (white asterisks) and euchromatin (red asterisks).
1. Nucleosomes (10 nm)
DNA is always associated to histones. Histones and DNA form nucleosomes, the basic unit of the chromatin organization (Figure 2). It can be said that nucleosomes are the more basic level of DNA packaging. In human cells, it is estimated that there are around 3,3x107 nucleosomes. A nucleosome is composed of a core of histones around which DNA wraps 1,65 times. There are around 166 base pairs (DNA is double strand) in each nucleosome. Two consecutive nucleosome cores are connected by linker DNA of around 34 base pairs. Thus, there are units of about 200 base pairs with are repeated along the DNA strand. However, the linker DNA may vary in length depending on the species or tissues, even between different regions of the same cell nucleus. The nucleosome core is formed by pairs of histones H3, H4, H2A and H2B, that is 8 histones. Each of these histones has a 30 amino acid sequence in the N terminal that protrudes from the protein. This sequence is highly conserved during evolution. It is like a tail with two main functions: regulates the access of other proteins to the DNA for transcription or repairing, and they also allows a higher level of chromatin condensation by interacting with neighbor nucleosomes.
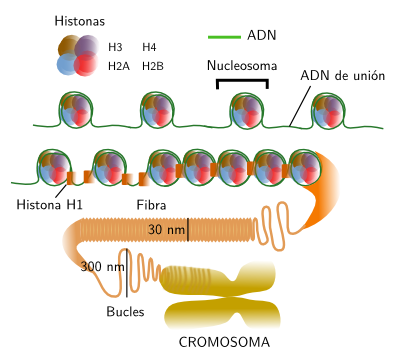
Figure 2. Organization levels of chromatin, from nucleosomes to chromosomes.
2. Fibers (30 nm)
H1 histone, or connection histone, is associated with the linker in a place close to the DNA exit from the nucleosome or to the entrance into the nucleosome. In mammals, there are at least 8 variants of H1. One of the roles of H1, together with nucleosome histons, is helping with the condensing of chromatin in 30 nm thick fibers. There are several models trying to explain how fibers are condensed into chromosomes, the highest compaction level of chromatin. Helix, zigzag and crossed linking organization have been suggested. Most authors propose that fibers are arranged in loops of about 300 nm. Those loops may be packaged in higher order structures called cromomeres (300-700 nm). Cromomeres could be present as dark aggregates in the heterechromatin of the interphase nucleus. However, many authors suggest that loops may be packed directly into chromosomes, without intermediary organizations. Heterochromatin may show different packaging levels of 30 nm fibers.
3. Chromosomes
The beginning of M phase means that both euchromatin and heterochromatin become chromosomes. This condensation is orchestrated by many proteins, including cohesins, condensins and toposiomerases. Each metaphase chromosome are composed of two sister chromatids synthesized during S phase. Initially, condensins keep attached the sister chromatids along the whole chromatid extension, but only by the centromeric region in later stages of M phase.
Many animal species are diploids, that is, their cells have two copie of each chromosome (each pair is composed of two homologous chromosomes, one from the mother and the other from the father) and therefore each gen has two copies which are known as alleles. A karyotype is the complete set of chromosomes of a species as they are in metaphase (Figure 3). The number, size and shape of chromosomes are characteristic of each species, although there are some exceptions. Karyotypes have two main types of chromosomes: autosomes and sexual chromosomes. Auotosomes are similar in males and females, but sexual chromosomes may different. For example, mammalian females have two X chromosomes whereas males have an X and a Y sexual chromosomes (the last couple of the first row in Figure 3). The number of chromosomes vary depending on the species, from 1 or 2 in some ant species, to more than 700 in some ferns.
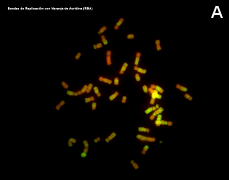

Figure 3. Metaphase chromosomes (click in each image to enlarge). Syrian hamster chromosomes obtained from metaphase cells and stained with acridine orange. Chromosome stained bands are replication bands. A: Picture of chromosomes as they are obtained onto the slide. B: Karyotype, that is, homologous chromosomes are paired and ordered (Images by Concepción Pérez García and Juan José Pasantes Ludeña. Dpt. Biochemistry, Genetic and Immunology. Faculty of Sciences. University of Vigo).
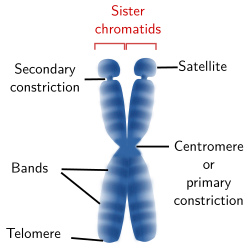
Figure 4. Drawing of a chromosome showing the structures that can be visualized at light microscopy. A chromosome may not show all these structures.
A light microscopy, several chromosome structures can be observed (Figure 4): centromere or primary constriction, secondary constriction, satellites, telomeres, and bands. The tips of the sister chromatides are called telomeres and the region where both chromatids are attached to each other is known as centromere or primary constriction.
Chromosome morphology is a major feature when making karyotypes because chromosomes can be compared and classified (Figures 3 and 5). The shape is greatly determined by the position of the centromere, because it influence the length of the chromosome arms, which are not usually equal. Metacentric chromosomes show arms with the similar length (for example A5 and E20 pairs in Figure 3). Submetacentric chromosomes have an arm clearly larger that the other (for example, A2 y C14 in Figure 3). Subtelocentric or acrocentric chromosomes have arms with different lengths, and this difference is higher than in metacentric chromosomsomes (see B6 y D19 in Figure 3). Telocentric chromosomes do not show one the arms, which means that chromatids are attatched between each other through their tips (see D16 and D17 in Figure 3).
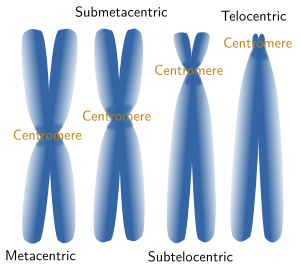
Figure 5. Chormosomes are named according to their morphology.
A typical centromere appears as a constriction, known as primary constriction, visible at light microscopy in metaphase chromosomes of higher eukaryotes (Figure 5). A primary constriction can be defined as a region of the chromosome where sister chromatids can not be distinguished between each other, being both chromatids thinner than in the other regions of the chromsome. Centromeres are specialized region made up of less condensed chromatin and are involved in the correct segregation of the chromosomes during anaphase. The kinetochore is a protein complex assembled at the centromere that gathers mitotic spindle microtubules, leading to a proper line up of chromosomes during metaphase and segregation during anaphase. There are diffuse and localized kinetochores. Localized kinetochores, the most frequent, are found at the centromeres and attach mitotic microtubules. Sometimes, kinetochore proteins are so concentrated that they link just one microtubule, like in some yeast species. Diffuse kinetochores, less frequent, have proteins scattered over the hole chromosome, so microtubules are attached along the whole chromosome.
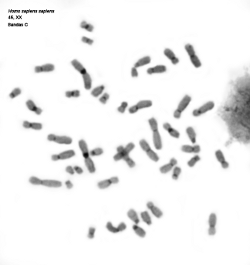
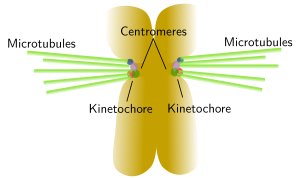
Figure 6. The image on the left shows human chromosomes with C bands or constitutive heterochromatine regions that are usually located close to centromeres. Kinetochores are assembled around centromeres and many mitotic spindle microtubules are linked to these regions, which is depicted in the image on the right. Click on the C bands image to enlarge. (Images by Concepción Pérez García and Juan José Pasantes Ludeña. Dpt. Biochemistry, Genetic and Immunology. Faculty of Sciences. University of Vigo).
Secondary constrictions are found in the arms of some chromosomes. These are regions with more loose chromatin, but without centromere. The best known secondary constriction is produced by chromatin regions that form the nucleolus, known as nucleolar organizing regions (NOR). Depending on the species, only one or a few chromosomes of the karyotype show this secondary constriction, which is found in the intermediate region (not terminal) of the chromosome arms. When the secondary constriction is located near the telomere, the chromosome segment between the telomere and the secondary constriction is known as satellite (Figure 4). In humans, the short arms of the human chromosomes 13, 14, 15, 21 and 22 show satellites.
At light microscopy, some special staining protocols and dyes can label bands with a variety of intensities or with different colors, some of them are fluorescentdyes (Figures 3 and 7). Band pattern is influenced by the chromosome pre-treatment and the staining protocol used. Structural (cytosine and guanine proportions, chromatin compaction) and functional (replication time) features of the DNA that form those bands also affect the visualization of bands. The band pattern is particular for each chromosome so that it is used for distinguishing chromosomes that have similar size and morphology. Band patterns are also very useful for detecting chromosome alterations or for accurately locating the physical position of certain genes in the chromosome.
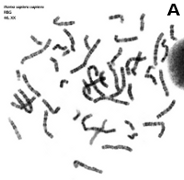
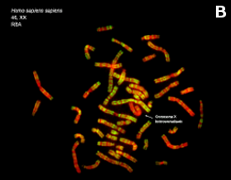
Figure 7. Metaphase chromosomes (click on the images to enlarge). A) Human chromosomes stained with Giemsa. B) Human chromosomes stained with acridine orange, a fluorescent dye (Images by Concepción Pérez García and Juan José Pasantes Ludeña. Dpt. Biochemistry, Genetic and Immunology. Faculty of Sciences. University of Vigo).
4. Decondensed chromosomes
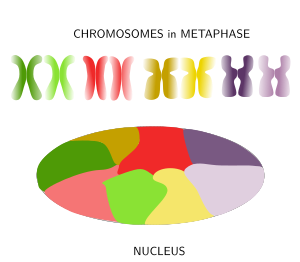
Figure 8. Chromosomes are decondensed during anaphase and chromatin is distributed in the nucleus in a tidy maner because each chromosome spread its chromatin into a defined territory that do not overlap much with the territories of other chromosomes. (Adapted from Bolzer et al., 2005).
When thinking of the nucleus in interphase with its huge among of chromatin after chromosome decondensation, it is difficult to imagine how cells can manage to condensate, decondensate, regulate and express the DNA without ending up in a monumental tangle. Actually, far from being a mess, it was observed in 1980 that in mammals the chromatin of each chromosome occupies a defined spatial territory (Figure 8). These territories are nearly spherical, 2 to 4 µm in diameter, and neighbor territories have little overlapping at their borders (however, in yeasts, the limits of these territories are not so well-defined). Many evidences support a nonrandom distribution of chromosome territories in the nucleus. For example, while territories disposition may change from cell to cell, during differentiation periods, and along the cell cycle, it looks like that the general pattern is quite stable in a cell during long time periods. The more active territories are usually found in the center of the nucleus, whereas the less active localize near the nuclear envelope. In a territory, regions that were replicated early during S phase (early replication regions) are segregated from those regions replicated during the second part of the S phase (late replication regions). The physical interactions of chromatin with the nuclear lamina and inner membrane of the nuclear envelope help with the segregation of chromosome territories. It is of notice that homologous chromosome also occupy different territories in the nucleus.
There are sub-territories or domains withing each territory (Figure 9). For example, there are repressed chromatin domains: policombic, heterochromatin and other less characterized heterochromatin organizations. There is also active or open chromatin that may contain regulatory sequences, promoters, transcribed sequences and regions linked to insulating proteins. Repressed chromatin of one territory does not interact with other domains, whereas open chromatin may do it, even with domains of other chromosome territories. Local interactions between chromatin loops may account for the existence of territory domains. For example, active chromatin is prone to interact with other nearby domains having active chromatin instead of repressed chromatin.
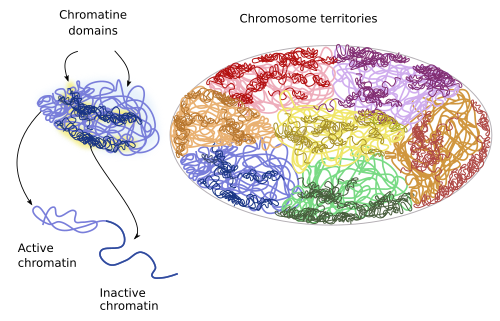
Figure 9. During interphase, chromatin territories include subterritories or domains.
Bibliografía
Belmond AS. Mitotic chromosome scaffold structure: new approaches to an old controversy. Proceedings of the National Academy of Sciences USA. 2002. 99:15855 -15857.
Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLOS biology. 2005. 3(5):e157. doi:10.1371/journal.pbio.0030157
Cavalli G, Misteli T. 2012. Functional implications of genome topology. Nature structural and molecular biology. 20: 290-299.
Vagnarelli P, Ribeiro SA, Earnshaw WC. Centromeres: old tales and new tools. FEBS Letters. 2008. 582:1950 -1959.
Wanner G, Formanek H. A new chromosome model. Journal of structural biology. 2000. 132:147 -161.